Applications of Sponge Iron and Effects of Organic Carbon Source on Sulfate-Reducing Ammonium Oxidation Process(1)
1. Introduction
In order to control the negative impact of climate change, many countries have reduced the emissions of greenhouse gases (GHG). China plays a crucial role in resisting climate change and has promised to reach its peak total GHG emissions by 2030. Although carbon dioxide (CO2) constitutes most of the greenhouse gases, other gases such as nitrous oxide (N2O) cause a physical effect known as radiative forcing (RF), which is the main driving force behind climate change. Nitrogen removal from wastewater chiefly relies on biological processes: nitrification and denitrification. Either nitrification or denitrification inevitably produces N2O, the greenhouse effect of which is 265 times higher than CO2. Current estimates consider that humans cause 40% of N2O emissions, and global wastewater is the fifth largest contributor to N2O emissions and continuously increases as the population grows and the industry develops.
The typical sulfate-reducing ammonium oxidation (SRAO) is an environmentally friendly denitrification process that converts ammonium (as the electron donor) and sulfate (as the electron acceptor) into nitrogen in an anaerobic environment, as expressed in Equation (1) :
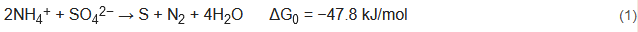
As autotrophic bacteria, anaerobic ammonia-oxidizing (anammox) bacteria do not generate other greenhouse gases in the anaerobic ammonium oxidation. However, the growth rate of the anammox bacteria is slow, and their nitrogen removal performance is unstable. Some studies have demonstrated that adding a 30% filling degree of zerovalent iron (ZVI) in the reactor strengthens their growth rate and stability. Sponge iron is a raw material for steel making, which is made from reducing iron ore to metallic iron, easy to obtain, and inexpensive. Sponge iron is a suitable carrier for nitrogen removal and enhances biological nitrogen removal. Moreover, it is a standard filler, which has the characteristics of a large specific surface area, a substantial reduction ability, and a loose and porous internal structure. Iron scraps can also stimulate both catabolism and anabolism in the presence of anaerobes.
Multiple factors, especially carbon sources, influence the coupling of ammonium oxidation and sulfur recycling. Various organic materials have different effects on the system, such as affecting sulfate reduction, denitrification, and other processes. Further, the concentration of the carbon source significantly impacts nitrogen removal. It has also been found that under organic conditions, a chemical oxygen demand (COD) of 160–5400 mg/L could improve heterotrophic processes, such as sulfate reduction and denitrification, thereby promoting the denitrification process. A deep understanding of the influences of the species and quantities of the carbon source is essential for the nitrogen and sulfur removal processes in SRAO to enhance some biological reactions and the efficiency of nitrogen removal.
Glucose, representing fatty acids, is a well-utilized organic carbon source for sulfate-reducing bacteria (SRB) that are heterotrophic. As an incompletely oxidized carbon source, sodium acetate promotes the growth of both sulfate-reducing bacteria and denitrifying bacteria. Phenol is chosen as the last group of organic carbon sources since it is a poisonous organic substance and has an inhibitory effect on many microorganisms; however, it is uncertain whether phenol has a negative impact on the SRAO and whether this effect is reversible. It is reported that when the concentration of the organic matter exceeds 300 mg/L, the activity of anammox declines due to the competitive inhibition between heterotrophic and autotrophic bacteria in the SRAO system. Therefore, the prerequisite to ensuring the effectiveness of nitrogen removal is to control the concentration and type of organic matter in the process.
Therefore, we try to address the challenges of the practical application of SRAO by two means. First, this study develops a new method to shorten the start-up time by adding sponge iron. Second, previous studies have not been conducted on how specific carbon source types affect the SRAO system; thus, this work explores new perspectives by looking at the changes in contaminants with the addition of different organic substances to the system and trying to explain them. This paper employs 4.8 kg of sponge iron in a 2.0 dm3 anaerobic sequencing batch reactor to enhance the start-up stage and the simultaneous removal of nitrogen and sulfate. Three organics, namely phenol, sodium acetate, and glucose, were chosen as the typical organic sources in the range of 75–200 mg/L and added into the sludge-seeded serum bottles to investigate the effect of different organics on the SRAO process. Moreover, it aims to explain the cooperative relationships of the various bacterial species by examining the microbial community structure and investigating the influence of the organic carbon source on the microbial community by the relative abundance at a genus level. This work also supplements the application of the SRAO process to convert industrial wastewater containing nitrogen and sulfur elements and provides a reference for a similar range of applications containing anaerobic ammonia oxidation processes.
2. Materials and Methods
2.1. Sponge Iron Treatment and Synthetic Wastewater
The sponge iron was purchased from Henan Zhengjie Environmental Protection Material Company (Zhengzhou, China). The materials were sieved to collect the fractions with a particle size of 1–2 mm. The sponge iron was soaked first in a 0.5% NaOH solution for 1 h to remove the organics on its surface and then in 1 mol/L of HCl for 0.5 h to remove inorganic impurities and metal oxides on its surface. Afterward, the treated sponge iron was cleaned with distilled water until the pH of the cleaning effluent was neutral. Finally, it was rinsed with absolute ethanol three times, dried in a ventilation system, sealed, and kept for further use. To expand the specific surface area of the sponge iron as much as possible, we chose particles with a size in the range of 1–2 mm. On the basis of the reactor specifications previously reported, the volume of the sponge iron added to each reactor was 0.6 dm3 ; thus, the mass of the sponge iron was 4.8 kg. Inoculated sludge was added into reactors ASBR1 and ASBR2, but the sponge iron was only added into reactor ASBR2.
The synthetic wastewater contained no nitrite but did contain sulfate to donate and accept electrons. Table 1 describes the composition of the synthetic wastewater. We used NH4Cl and K2SO4 as the primary influent substances and controlled NH4+ and SO42− in the range of 25.2–42.1 and 186.7–311.1 mg/L, respectively. The different carbon sources, namely phenol, sodium acetate, and glucose, were added into anaerobic serum bottles A1, A2, and A3, respectively. The chemical oxygen demand also ranged from 167 to 447 mg/L.
Table 1. The composition of the synthetic wastewater.
| Chemicals | Concentration (mg/L) |
| KH2PO4 | 27 |
| CaCl2 | 180 |
| MgSO4·7H2O | 300 |
| NaHCO3 | 1250 |
| NH4Cl | 125 |
| K2SO4 | 444 |
| H3BO3 | 0.14 |
| MnCl2·4H2O | 9.9 |
| CuSO4·5H2O | 2.5 |
| ZnSO4·7H2O | 4.3 |
| NiCl2·6H2O | 1.9 |
| Na2MoO4·2H2O | 2.2 |
| CoCl2·6 H2O | 24 |
| EDTA | 5 |
| FeSO4 | 5 |
2.2. Reactor Setup and Operation
Anaerobic sequencing batch reactors were employed to culture the bacteria for SRAO and investigate the effect of the sponge iron. The sludge used in the experiments was composed of the conventional anaerobic ammonium oxidation sludge preserved by the research group and the sludge from the anaerobic section of the Changchun Southeast sewage treatment plant (China). Seed sludge (0.5 L) was added into each ASBR, resulting in an initial value of the mixed liquor suspended solids (MLSS) equal to 2200 mg/L. The experiments were conducted using two sets of laboratory-scale ASBRs for sulfate-type anaerobic ammonium oxidation during the start-up process, as shown in Figure 1. The experimental setup was cylindrical with an adequate volume of 2.0 dm3. The reactor was equipped with an external, constant-temperature water jacket, and the water jacket compartment was 2 cm-thick and wrapped in black insulation cotton, which could protect the reactor from light and reduce heat loss to maintain a constant temperature. To control the dissolved oxygen (DO) content, we purged the influent containing the target elements by nitrogen gas before entering the reactor, and a dissolved oxygen meter measured the DO content. The ASBR reactor was operated in 48 h cycles consisting of 4 phases: an influent phase (single influent for 1 h), a reaction phase (intermittent stirring for 24 h), a sedimentation phase (22 h), and a drainage phase (1 h). The reactor received water through a peristaltic pump, and each reactor was equipped with a mechanical stirrer with a controllable speed. The stirring duration was 24 h per cycle, and the rotation speed of the stirrer was 150 rpm. Further, the whole process operated autonomously through a program controller, and the reactor temperature was set at 35 °C using the water jacket. A volume of 1.0 dm3 of the supernatant drained per cycle was taken as the testing sample, and new synthetic wastewater was added to the system.
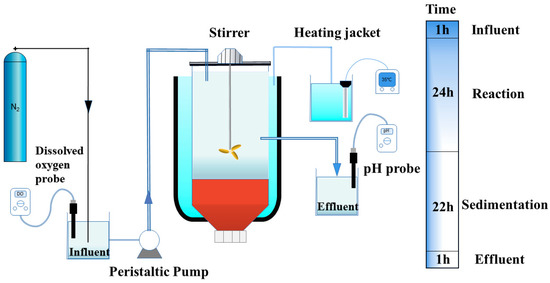
Figure 1. The anaerobic sequencing batch reactor and the operation time in a cycle.
2.3. Sequencing Batch Experiments
The modified anaerobic serum bottle method was used in this work. To this end, three 250 mL serum bottles were firstly sterilized, and their neck was plugged with a butyl rubber stopper for ready use. After 90 days of cultivation, the ASBR was successfully started up, and the effect of nitrogen and sulfur removal was stabilized. Each time, 200 mL of the mixture was removed from the ASBR and subjected to centrifugation at a speed of 4000 rpm for 10 min under anaerobic conditions. Next, the supernatant was removed, and the retentate was washed with an equal volume of the nutrient stock solution, followed by a second centrifugation to turn the culture into a suspension. The substrate was then added to a 250 mL serum bottle with the target components. The bottles containing phenol, sodium acetate, and glucose were marked A1, A2, and A3, respectively, depending on the added organic substance. Like the ASBR reactor, after each addition of synthetic wastewater, the serum bottle was shaken in the shaker at 150 rpm for 24 h, and the supernatant was discharged after 22 h of sedimentation in one cycle. The environmental conditions were consistent with the ASBR, and 100 mL of the supernatant was extracted with a medical syringe for testing every 48 h.
2.4. Measurements
The various organic masses in water were uniformly measured according to the measurement method of COD. The standard method also measured COD, NH4+, SO42−, NO2−, and NO3. The mixed liquor suspended solids and the mixed liquid volatile suspended solids (MLVSS) were determined gravimetrically. The above analytical methods were derived from the fourth edition of the Water and Wastewater Monitoring and Analysis Method. The TN was also calculated as the sum of the concentrations of nitrate, nitrite, and ammonium nitrogen. The dissolved oxygen and pH were measured manually by a dissolved oxygen meter (HQ30D, Hach, Loveland, CO, USA) and a pH meter (Phs-25, LiDa, Shanghai, China), respectively. The ultraviolet-visible (UV-vis) spectroscopy was conducted by a spectrophotometer model UV2400 (Hengping, Shanghai, China). Origin 8.5 performed data plotting and statistical analysis.
2.5. Analysis of Microbial Community
The microbial community structures of the tested systems were analyzed via the Illumina high-throughput sequencing technology. To this end, the sludge samples were taken from the three anaerobic serum bottles of phenol, sodium acetate, and glucose at a period of 60 days and marked A1, A2, and A3, respectively. The total genomic DNA was also extracted using a PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA). Bacterial 16S rRNA gene fragments were amplified via the polymerase chain reaction (PCR) with the primer set 338F (ACTCCTACGGGAGGCAGCAG)/806R (GGACTACHVGGGTWTCTAAT). Majorbio Biotech Co., Ltd., Shanghai, China, analyzed the microbial community structures. The data from the measurement results were statistically analyzed using the SPSSAU platform (Qing Si Technology Ltd, Beijing, China) and computational analysis.




